Biography
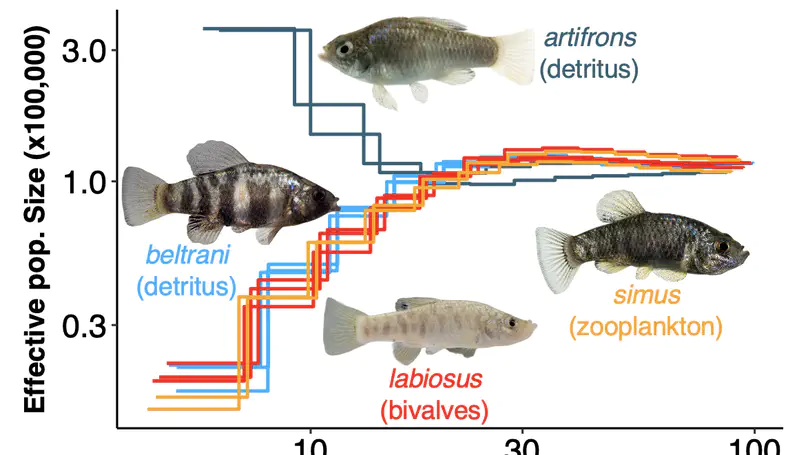
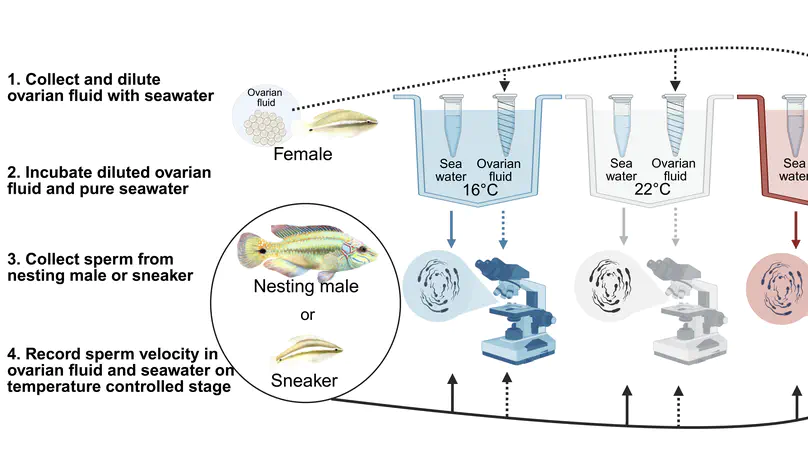
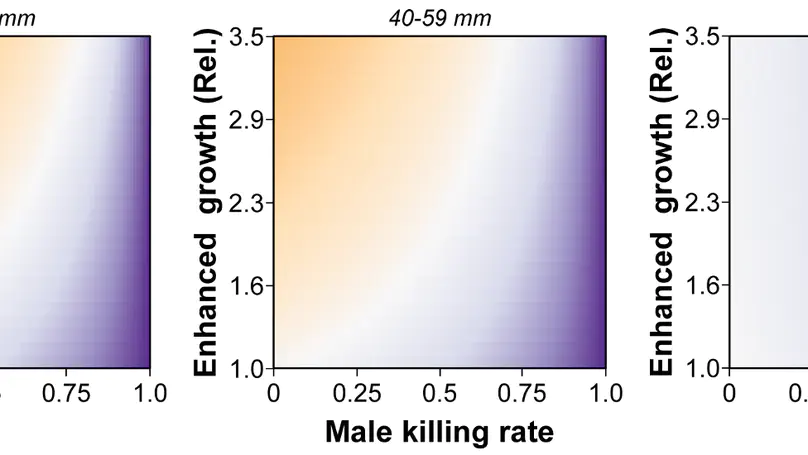
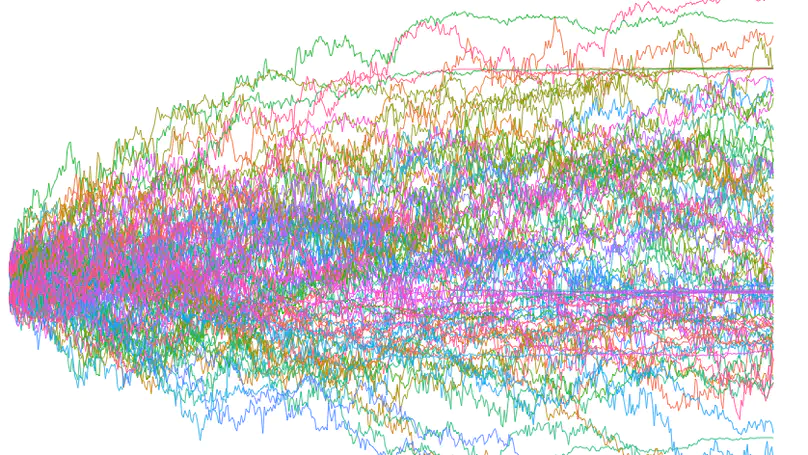
My research integrates fieldwork, behavioral, experimental, genomic, and theoretical approaches to understand the ecology and evolution of reproductive interactions. Overarching questions that guide my research program are: (1) How do biotic and abiotic environments interact to influence reproductive interactions, (2) how does this affect the evolution of life-history traits and speciation, and (3) what is the underlying genetic architecture of traits involved in these reproductive interactions? I am currently a Miller Research Fellow at UC Berkeley in Dr. Christopher Martin’s lab. As a Miller Fellow, I am exploring how natural and sexual selection interact to rapidly create many new species by developing new theoretical models and testing them with empirical work on pupfish. My PhD work was with Dr. Suzanne Alonzo at the University of California, Santa Cruz. For my dissertation, I researched how females influence the evolution of male behavior via cryptic female choice (females bias fertilization to specific males), how climate change influences female-sperm interactions, and the role that cryptic female choice might play in speciation. I also develop mathematical models to understand the ecological-evolutionary dynamics of microbe-host interactions in marine invertebrates.
Download my CV.
Any opinion, findings, and conclusions or recommendations expressed in material on this website are those of my own.
- Coevolution
- Speciation
- Sexual Selection
- Alternative Reproductive tactics
- R
- Julia
-
PhD Ecology and Evolutionary Biology, 2024
University of California, Santa Cruz
-
B.S in Biology and B.A. Computer Science, 2018
University of Virginia